Analysis Tutorials
Here is a collection of tutorials specifically focused on performing
analysis in Amber.
Back to main tutorial page.
CPPTRAJ
PYTRAJ
CPPTRAJ tutorials
These tutorials focus on analyzing MD simulations with CPPTRAJ.
TUTORIAL C0: An Introduction to CPPTRAJ
 This tutorial will give a basic introduction to using CPPTRAJ for performing
trajectory analysis. It will cover using CPPTRAJ interactively and in batch
mode for processing scripts, loading topologies and trajectories,
processing data, and working with data sets.
This tutorial will give a basic introduction to using CPPTRAJ for performing
trajectory analysis. It will cover using CPPTRAJ interactively and in batch
mode for processing scripts, loading topologies and trajectories,
processing data, and working with data sets.
By Daniel R. Roe
TUTORIAL C1: RMSD Analysis in CPPTRAJ
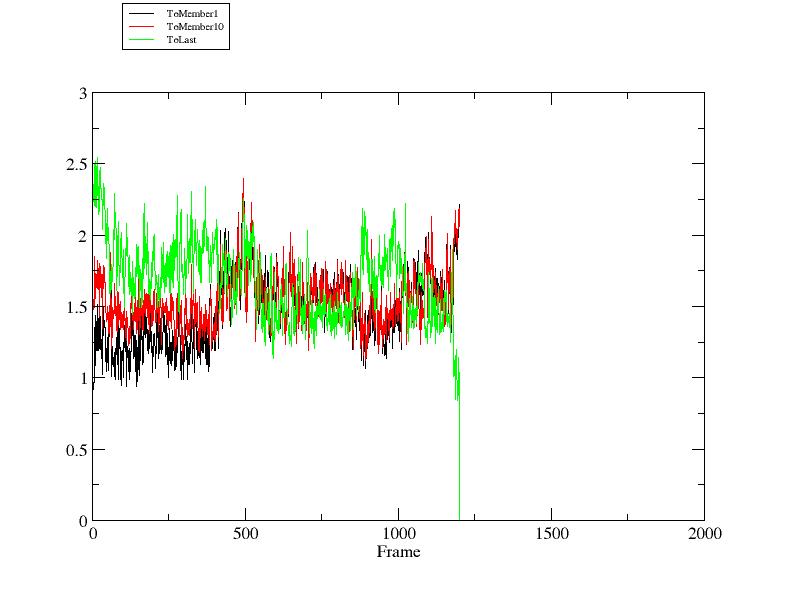 This tutorial will give a basic introduction to performing RMSD calculations
with CPPTRAJ. It will cover loading reference structures, as well as calculating
RMSD to references with different topologies.
This tutorial will give a basic introduction to performing RMSD calculations
with CPPTRAJ. It will cover loading reference structures, as well as calculating
RMSD to references with different topologies.
By Daniel R. Roe
TUTORAL C4: Combined Clustering Analysis with CPPTRAJ
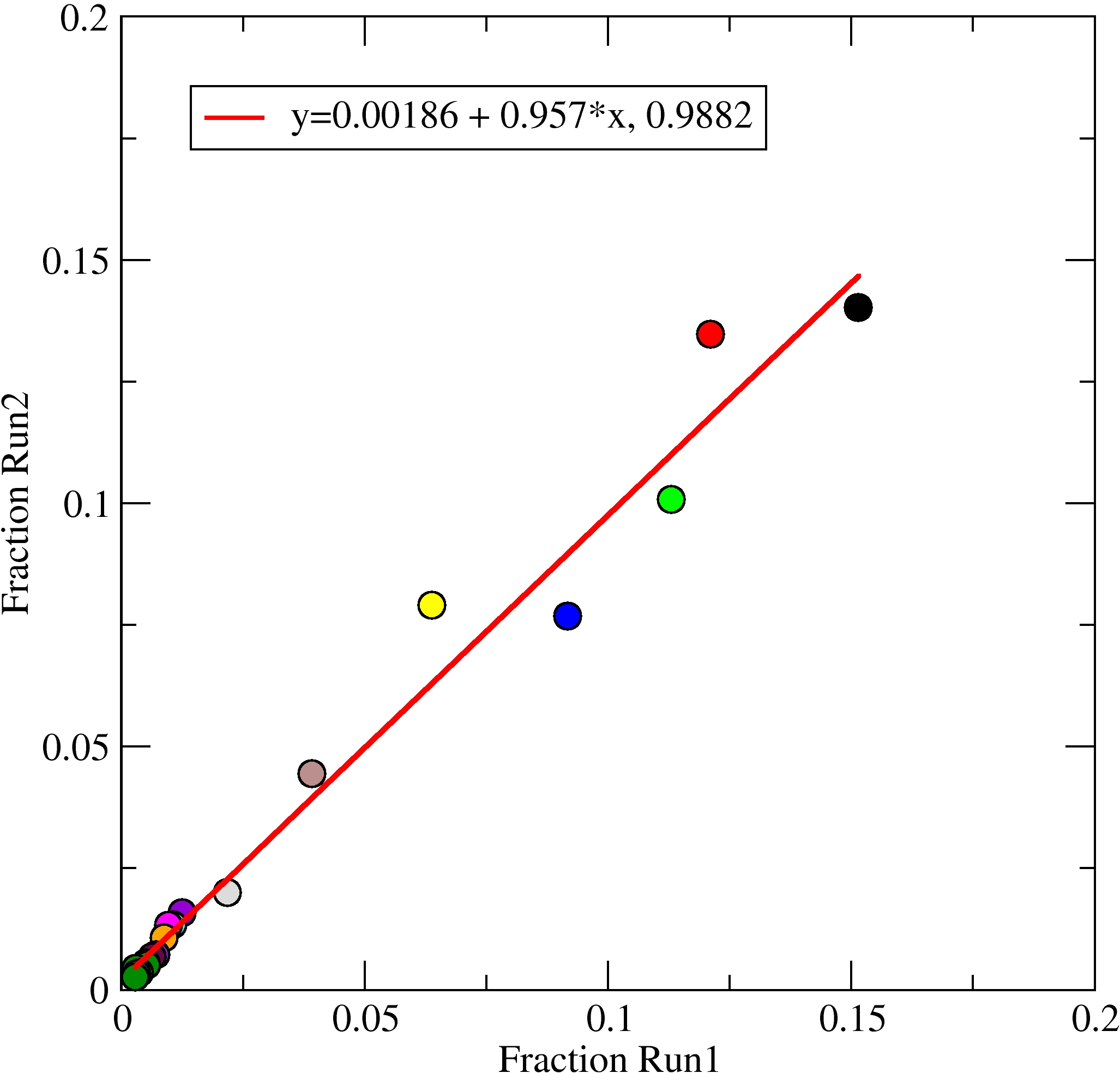 This tutorial will cover how to perform combined clustering analysis with
CPPTRAJ, which is a way of comparing structure populations between two
or more independent trajectories or between different parts of a single
trajectory.
This tutorial will cover how to perform combined clustering analysis with
CPPTRAJ, which is a way of comparing structure populations between two
or more independent trajectories or between different parts of a single
trajectory.
By Daniel R. Roe
PYTRAJ tutorials
These tutorials focus on analyzing and visualizing MD simulations with PYTRAJ and
NGLView.
Back to main tutorial page.
 This tutorial will give a basic introduction to using CPPTRAJ for performing
trajectory analysis. It will cover using CPPTRAJ interactively and in batch
mode for processing scripts, loading topologies and trajectories,
processing data, and working with data sets.
This tutorial will give a basic introduction to using CPPTRAJ for performing
trajectory analysis. It will cover using CPPTRAJ interactively and in batch
mode for processing scripts, loading topologies and trajectories,
processing data, and working with data sets.