AMBER 12 NVIDIA GPU
ACCELERATION SUPPORT
THIS IS AN ARCHIVED PAGE FOR AMBER
v12 GPU SUPPORT
IT IS NOT ACTIVELY BEING
UPDATED
FOR THE LATEST VERSION OF THESE
PAGES PLEASE
GO HERE
Benchmarks
Benchmarks timings by Mike Wu and Ross
Walker.
Download AMBER
Benchmark Suite
Please Note: The current benchmark
timings here are for AMBER 12 up to and including Bugfix.19 (GPU support revision 12.3.1,
Aug 15th 2013).
Machine Specs
Machine 1
CPU = Dual x 6 Core Intel E5-2640 (2.5GHz)
MPICH2 v1.5 - GNU v4.4.7-3
GPU = GTX580 (1.5GB) / GTX680 (4.0GB) / GTX770 (4.0GB) / GTX780
(3.0GB) / GTX-Titan (6.0GB)
nvcc v5.0
NVIDIA Driver Linux 64 - 325.15
Machine 2
CPU = Dual x 8 Core Intel E5-2687W @ 3.10 GHz
Motherboard = SuperMicro X9DR3-F Motherboard
GPU = K10 (2x4GB) / K20 (5GB) / K20X (6GB) / K40 (12GB)
ECC = OFF
nvcc v5.0
NVIDIA Driver Linux 64 - 304.51
Machine
3 (SDSC Gordon)
CPU = Dual x 8 Core Intel E5-2670 @ 2.60GHz
MVAPICH2 v1.8a1p1
Intel Compilers v12.1.0
QDR IB Interconnect
K10 Note: The K10 naming is a little
confusing. In these plots we have chosen to refer to K10's as the
number of GPUs exposed to the operating system. Thus 2 x K10 is
actually a single K10 card and 8 x K10 means 4 K10 cards.
Code Base = AMBER 12 Release + Bugfixes 1
to 19
- GPU code v12.3.1 (Aug 2013)
Precision Model = SPFP (GPU), Double Precision
(CPU)
Benchmarks were run with ECC turned OFF on
GTX/M2090/K10/K20/K40 cards - we have seen no issues with
AMBER reliability related to ECC being on or off. If you see approximately 10% less
performance than the numbers here then run the following (for each GPU) as root:
nvidia-smi -g 0
--ecc-config=0 (repeat
with -g x for each GPU ID)
Boost Clocks: Some of the latest NVIDIA
GPUs, such as the K40, support boost clocks which increase the clock
speed if power and temperature headroom is available. This should be
turned on as follows to enable optimum performance with AMBER:
sudo nvidia-smi -i 0 -ac 3004,875
which puts device 0 into the highest boost state.
To return to normal do: sudo
nvidia-smi -rac
To enable this setting without being root do:
nvidia-smi -acp 0
Segfaults in Parallel: If you find that
runs across multiple nodes (i.e. using the infiniband adapter)
segfault almost immediately then this is most likely an issue with
GPU Direct v2 (CUDA v4.2/5.0) not being properly supported by your
hardware and driver installations. In most cases setting the
following environment variable on all nodes (put it in your .bashrc)
will fix the problem:
export
CUDA_NIC_INTEROP=1
List of Benchmarks
Explicit Solvent (PME)
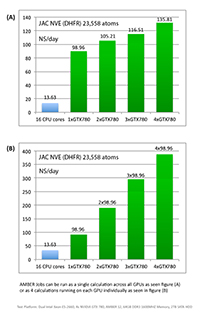
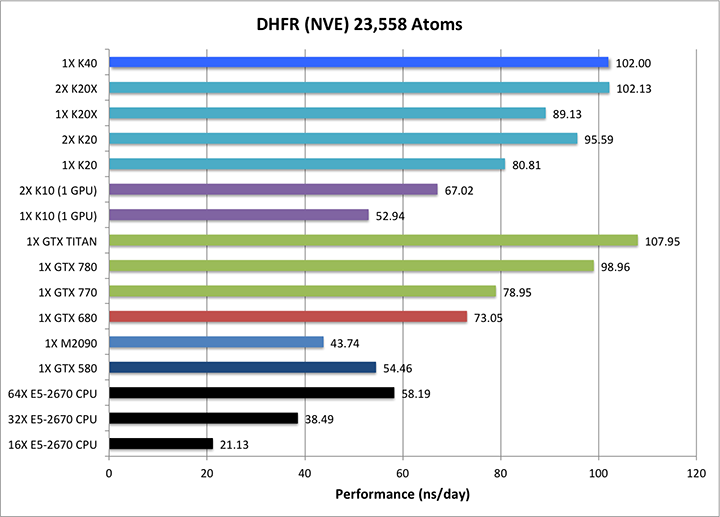
- DHFR NVE = 23,558 atoms
- DHFR NPT = 23,558 atoms
- FactorIX NVE = 90,906 atoms
- FactorIX NPT = 90,906 atoms
- Cellulose NVE = 408,609 atoms
- Cellulose NPT = 408,609 atoms
Implicit Solvent (GB)
- TRPCage = 304 atoms
- Myoglobin = 2,492 atoms
- Nucleosome = 25,095 atoms
You can download a tar file containing the input
files for all these benchmarks
here
(50.3 MB)
Individual vs Aggregate Performance
A unique feature of AMBER's GPU support that sets it apart from the
likes of Gromacs and NAMD is that it does NOT rely on the CPU to
enhance performance while running on a GPU. This allows one to make
extensive use of all of the GPUs in a multi-GPU node with maximum
efficiency. It also means one can purchase low cost CPUs making GPU
accelerated runs with AMBER substantially more cost effective than
similar runs with other GPU accelerated MD codes.
For example, suppose you have a node with 4
GTX-Titan GPUs in it. With a lot of other MD codes you can use one
to four of those GPUs, plus a bunch CPU cores for a single job.
However, the remaining GPUs are not available for additional jobs
without hurting the performance of the first job since the PCI-E bus
and CPU cores are already fully loaded. AMBER is different. During a
single GPU run the CPU and PCI-E bus are barely used. Thus you have
the choice of running a single MD run across multiple GPUs, to
maximize throughput on a single calculation, or alternatively you
could run four completely independent jobs one on each GPU. In this
case each individual run, unlike a lot of other GPU MD codes, will
run at full speed. For this reason AMBER's aggregate throughput on
cost effective multi-GPU needs massively exceeds that of other codes
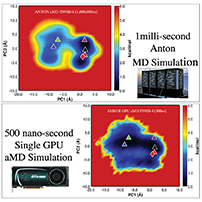
that rely on constant CPU to GPU communication. This is illustrated
below in the plots showing 'aggregate' throughput.
^
|


|
|
Explicit Solvent PME Benchmarks |
|
1) DHFR NVE = 23,558 atoms
Typical Production MD NVE with
GOOD energy conservation.
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2, tol=0.000001,
nstlim=10000,
ntpr=1000, ntwx=1000,
ntwr=10000,
dt=0.002, cut=8.,
ntt=0, ntb=1, ntp=0,
ioutfm=1,
/
&ewald
dsum_tol=0.000001,
/
|
|
|
Single job throughput
(a single run on one or more GPUs and one or more nodes)
|
|
Aggregate throughput (GTX-Titan)
(individual runs at the same time on the same node)
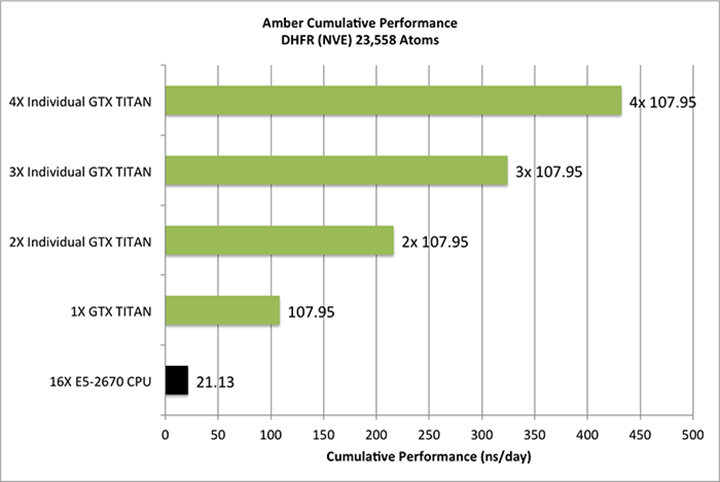
|
|
2) DHFR NPT = 23,558 atoms
Typical Production MD NPT
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2,
nstlim=10000,
ntpr=1000, ntwx=1000,
ntwr=10000,
dt=0.002, cut=8.,
ntt=1, tautp=10.0,
temp0=300.0,
ntb=2, ntp=1, taup=10.0,
ioutfm=1,
/
|

|
|
Single job throughput
(a single run on one or more GPUs and one or more nodes)
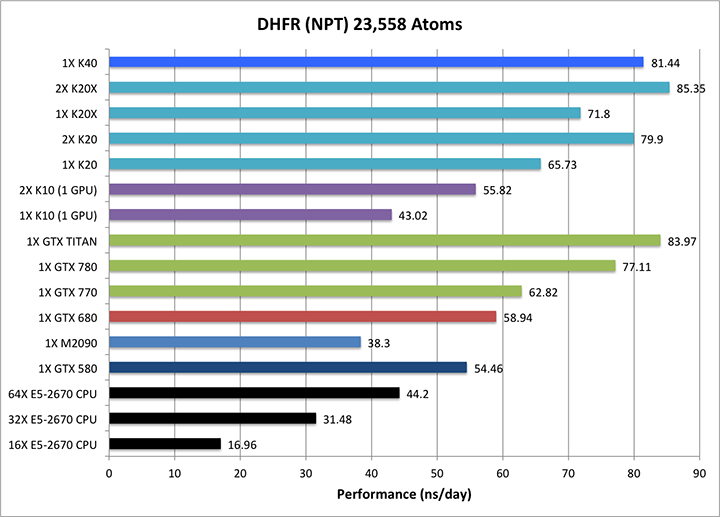
|
|
3) Factor IX NVE = 90,906 atoms
Typical Production MD NVE with
GOOD energy conservation.
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2, tol=0.000001,
nstlim=10000,
ntpr=1000, ntwx=1000,
ntwr=10000,
dt=0.002, cut=8.,
ntt=0, ntb=1, ntp=0,
ioutfm=1,
/
&ewald
dsum_tol=0.000001,nfft1=128,nfft2=64,nfft3=64,
/
|
 |
Single job throughput
(a single run on one or more GPUs and one or more nodes)
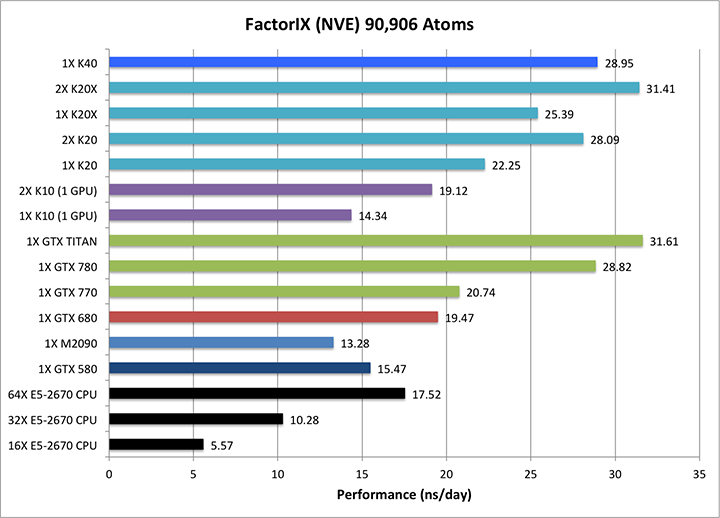
|
|
4) Factor IX NPT = 90,906 atoms
Typical Production MD NVT
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2,
nstlim=10000,
ntpr=1000, ntwx=1000,
ntwr=10000,
dt=0.002, cut=8.,
ntt=1, tautp=10.0,
temp0=300.0,
ntb=2, ntp=1, taup=10.0,
ioutfm=1,
/
|
 |
Single job throughput
(a single run on one or more GPUs and one or more nodes)
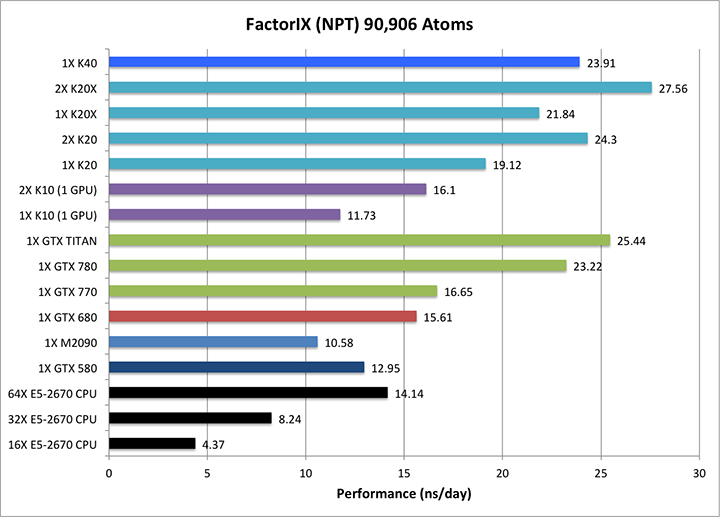
|
|
5) Cellulose NVE = 408,609 atoms
Typical Production MD NVE with
GOOD energy conservation.
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2, tol=0.000001,
nstlim=10000,
ntpr=1000, ntwx=1000,
ntwr=10000,
dt=0.002, cut=8.,
ntt=0, ntb=1, ntp=0,
ioutfm=1,
/
&ewald
dsum_tol=0.000001,
/
|
 |
Single job throughput
(a single run on one or more GPUs and one or more nodes)
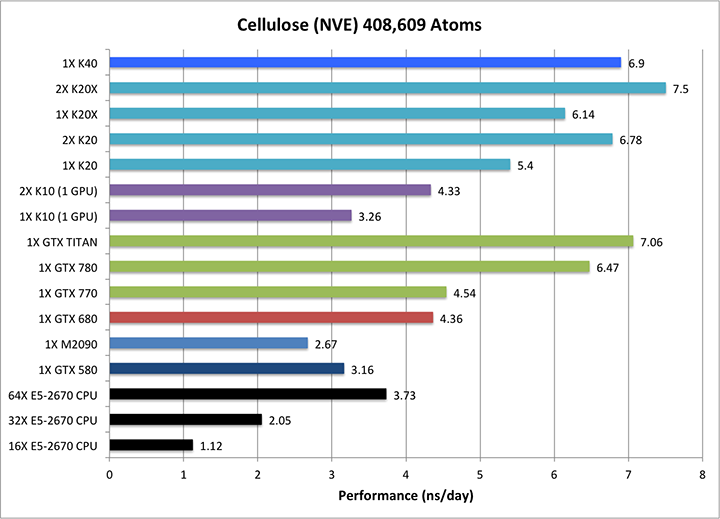
|
|
6) Cellulose NPT = 408,609 atoms
Typical Production MD NPT
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2,
nstlim=10000,
ntpr=1000, ntwx=1000,
ntwr=10000,
dt=0.002, cut=8.,
ntt=1, tautp=10.0,
temp0=300.0,
ntb=2, ntp=1, taup=10.0,
ioutfm=1,
/
|
 |
Single job throughput
(a single run on one or more GPUs and one or more nodes)
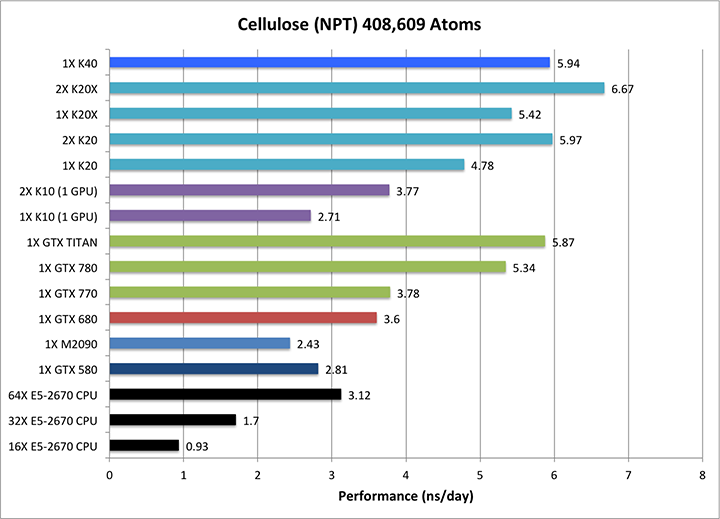
|
|
^
|
Implicit Solvent GB Benchmarks |
|
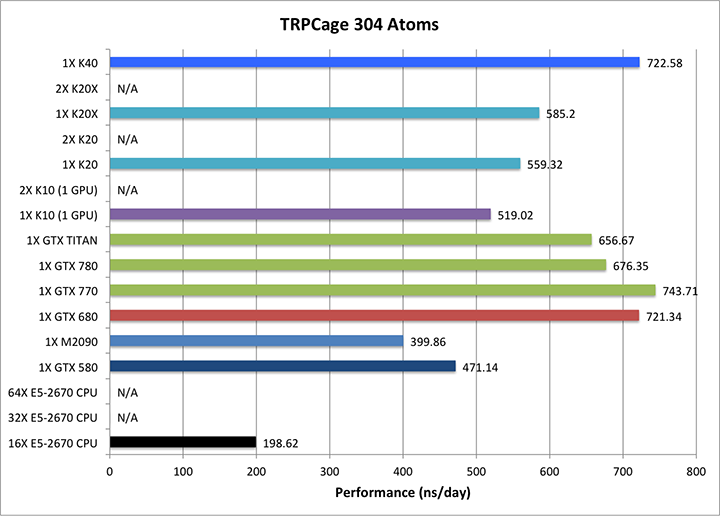
1) TRPCage = 304 atoms
&cntrl
imin=0,irest=1,ntx=5,
nstlim=100000,dt=0.002,ntb=0,
ntf=2,ntc=2,tol=0.000001,
ntpr=1000, ntwx=1000, ntwr=50000,
cut=9999.0, rgbmax=15.0,
igb=1,ntt=0,nscm=0,
/ Note: The TRPCage test is too small to make effective
use of the very latest GK110 (GTX780/Titan/K20) GPUs hence
performance on these cards is not as pronounced over early
generation cards as it is for larger GB systems and PME runs. |
 |
|
|
|
2) Myoglobin = 2,492 atoms
&cntrl
imin=0,irest=1,ntx=5,
nstlim=10000,dt=0.002,ntb=0,
ntf=2,ntc=2,tol=0.000001,
ntpr=1000, ntwx=1000,
ntwr=50000,
cut=9999.0, rgbmax=15.0,
igb=1,ntt=0,nscm=0,
/
|
 |
|

|
|
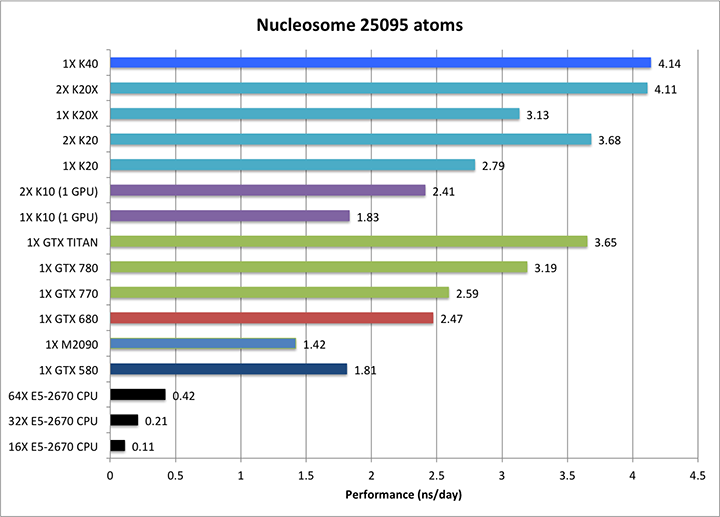
3) Nucleosome = 25095 atoms
&cntrl
imin=0,irest=1,ntx=5,
nstlim=1000,dt=0.002,ntb=0,
ntf=2,ntc=2,tol=0.000001,
ntpr=100, ntwx=100,
ntwr=50000,
cut=9999.0, rgbmax=15.0,
igb=1,ntt=0,nscm=0,
/
|
 |
|
|
^
|



















