Creating Special Systems for Molecular Simulations
TUTORIAL B5: Simulating the Green
Fluorescent Protein and building a modified amino acid residue
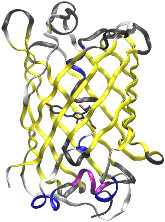 In this tutorial, we will learn how to
use the AMBER programs to build a residue template and parameter set for a
custom, modified amino acid. Unlike other tutorials that detail how to
create a residue template and parameter set for a small organic ligand, the
modified amino acid in this example must be bonded to the residues that come
before and after it in the protein polymer sequence. As a result, the
process is more complex and has more steps. In this tutorial, we will learn how to
use the AMBER programs to build a residue template and parameter set for a
custom, modified amino acid. Unlike other tutorials that detail how to
create a residue template and parameter set for a small organic ligand, the
modified amino acid in this example must be bonded to the residues that come
before and after it in the protein polymer sequence. As a result, the
process is more complex and has more steps.
By Jason Swails, Dave Case, and Taisung Lee
TUTORIAL A16: An Amber Lipid
Force Field Tutorial: Lipid14
 Updated for Lipid14: Phospholipid bilayers are essential components
to cellular membranes and are the stage where many essential biophysical and
biochemical processes take place. This tutorial explains how to set up and
simulate lipid bilayers with the Lipid14 force field. A DOPC bilayer is
built, converted, and loaded into LEaP to assign parameters for molecular
dynamics simulation. A molecular dynamics scheme is presented followed by
analysis of the bilayer structural properties in the trajectory.
Furthermore, membrane-bound proteins are examined and a simple membrane-bound
protein system is built.
Updated for Lipid14: Phospholipid bilayers are essential components
to cellular membranes and are the stage where many essential biophysical and
biochemical processes take place. This tutorial explains how to set up and
simulate lipid bilayers with the Lipid14 force field. A DOPC bilayer is
built, converted, and loaded into LEaP to assign parameters for molecular
dynamics simulation. A molecular dynamics scheme is presented followed by
analysis of the bilayer structural properties in the trajectory.
Furthermore, membrane-bound proteins are examined and a simple membrane-bound
protein system is built.
By Ben Madej and Ross Walker
Parker de Waal has created an
alternative lipid-building tutorial using Maestro.
| 

