(Note: These tutorials are meant to provide
illustrative examples of how to use the AMBER software suite to carry out
simulations that can be run on a simple workstation in a reasonable period of
time. They do not necessarily provide the optimal choice of parameters or
methods for the particular application area.)
Copyright Ross Walker 2010
Nudged Elastic Band (NEB) simulations - SECTION 6
Formerly known as TUTORIAL A5: The Nudged Elastic Band Approach to Finding the Lowest Energy Pathway Between two States
By Christina Bergonzo, Carlos Simmerling & Ross Walker
6. Slow Cooling
The final stage is to slowly cool the system down so that the images freeze out along the pathway. We will do the cooling in 2 stages. In the first stage we shall gradually cool down in 50 K steps over 120 ps using the following profile:
0 to 10ps : 300 K -> 250 K
10 to 20ps : Equil at 250 K
20 to 30ps : 250 K -> 200 K
30 to 40ps : Equil at 200 K
40 to 50ps : 200 -> 150 K
50 to 60ps : Equil at 150 K
60 to 70ps : 150 K -> 100 K
70 to 80ps : Equil at 100 K
80 to 90ps : 100 K -> 50 K
90 to 100ps : Equil at 50 K
100 to 110 ps : 50 K -> 0 K
110 to 120 ps : Equil at 0 K
Note since we are now back at 300 K and below we can safely switch back to using a 1 fs time step. Here is the script to create the input file and groupfile:
#!/bin/bash mkdir 4COOL1/ cd 4COOL1/ if [[ -e groupfile ]] ; then rm groupfile fi for ((i=0; i <= 31; i++)) ; do ext=`printf "%03i" $i` echo -O -p ../str1.prmtop -c ../3ANNEAL/neb.r.$ext -i ./4cool1.in -x ./neb.x.$ext -o ./neb.out.$ext \ -inf ./neb.info.$ext -r ./neb.r.$ext >> groupfile done cat > 4cool1.in <<EOF 120ps NEB ALA-ALA cool stage 1 &cntrl imin = 0, irest = 1, ntx=5, ntc=1, ntf=1, ntpr=500, ntwx=10000, ntb = 0, cut = 999.0, rgbmax=999.0, igb = 1, saltcon=0.2, nstlim = 120000, nscm= 0, dt = 0.001, ntt = 3, gamma_ln=1000.0, temp0=300, ineb = 1,skmin = 50,skmax = 50, tgtfitmask=":1,2,3", tgtrmsmask=":1,2,3@N,CA,C", nmropt=1,ig=-1, / &wt type='TEMP0', istep1=0,istep2=10000, value1=300.0, value2=250.0 / &wt type='TEMP0', istep1=10001,istep2=20000, value1=250.0, value2=250.0 / &wt type='TEMP0', istep1=20001,istep2=30000, value1=250.0, value2=200.0 / &wt type='TEMP0', istep1=30001,istep2=40000, value1=200.0, value2=200.0 / &wt type='TEMP0', istep1=40001,istep2=50000, value1=200.0, value2=150.0 / &wt type='TEMP0', istep1=50001,istep2=60000, value1=150.0, value2=150.0 / &wt type='TEMP0', istep1=60001,istep2=70000, value1=150.0, value2=100.0 / &wt type='TEMP0', istep1=70001,istep2=80000, value1=100.0, value2=100.0 / &wt type='TEMP0', istep1=80001,istep2=90000, value1=100.0, value2=50.0 / &wt type='TEMP0', istep1=90001,istep2=100000, value1=50.0, value2=50.0 / &wt type='TEMP0', istep1=100001,istep2=110000, value1=50.0, value2=0.0 / &wt type='TEMP0', istep1=110001,istep2=120000, value1=0.0, value2=0.0 / &wt type='END' / EOF cd .. |
The file creates a directory named 4COOL1, since this is the first cooling step. We can cd to this directory and run sander:
mpirun -np 32 $AMBERHOME/exe/sander.MPI -ng 32 -groupfile groupfile
This takes about 22 minutes, and produces the following output files: 4COOL1.tar.gz
Normally we would be done running simulated annealing at this point, however, the NEB method needs a final energy minimization of the path. This is done here using the velocity Verlet algorithm:
We will do one final 200ps long stage of cooling where we will set temp0=0.0 K and we will turn on the quenched velocity verlet method that was discussed in section 1 (vv=1). Here is the last script which creates an input file and groupfile for the final quenched MD at 0K:
#!/bin/bash mkdir 5COOL2/ cd 5COOL2/ if [[ -e groupfile ]] ; then rm groupfile fi for ((i=0; i <= 31; i++)) ; do ext=`printf "%03i" $i` echo -O -p ../str1.prmtop -c ../4COOL1/neb.r.$ext -i ./5cool2.in -x ./neb.x.$ext -o ./neb.out.$ext \ -inf ./neb.info.$ext -r ./neb.r.$ext >> groupfile done cat > 5cool2.in <<EOF 50ps NEB ALA-ALA cool stage 2 &cntrl imin = 0, irest = 1, ntx=5, ntc=1, ntf=1, ntpr=100, ntwx=10000, ntb = 0, cut = 999.0, rgbmax=999.0, igb = 1, saltcon=0.2, nstlim = 200000, nscm= 0, dt = 0.001, ntt = 3, gamma_ln=1000.0, temp0=0.0, ineb = 1,skmin = 50,skmax = 50, tgtfitmask=":1,2,3", tgtrmsmask=":1,2,3@N,CA,C", vv=1,vfac=0.1,ig=-1, / EOF cd .. |
We can run this as we do above:
mpirun -np 32 $AMBERHOME/exe/sander.MPI -ng 32 -groupfile groupfile
This takes about 40 minutes, and produces the following output files: 5COOL2.tar.gz
Lets take a look at the final step of this final cooling (looking at neb.out.003):
NSTEP = 200000 TIME(PS) = 740.000 TEMP(K) = 0.00 PRESS = 0.0 Etot = -36.1351 EKtot = 0.0000 EPtot = -36.1351 BOND = 0.5727 ANGLE = 1.3399 DIHED = 8.9864 1-4 NB = 2.9599 1-4 EEL = 44.7642 VDWAALS = -1.8926 EELEC = -75.0851 EGB = -17.7805 RESTRAINT = 0.0000 NEB replicate breakdown: Energy for replicate 1 = -36.0518 Energy for replicate 2 = -36.1171 Energy for replicate 3 = -36.1171 Energy for replicate 4 = -36.1351 Energy for replicate 5 = -36.1475 Energy for replicate 6 = -36.1528 Energy for replicate 7 = -36.1528 Energy for replicate 8 = -36.1529 Energy for replicate 9 = -36.1428 Energy for replicate 10 = -36.1130 Energy for replicate 11 = -36.1129 Energy for replicate 12 = -36.1129 Energy for replicate 13 = -36.0827 Energy for replicate 14 = -36.0490 Energy for replicate 15 = -36.0064 Energy for replicate 16 = -35.9555 Energy for replicate 17 = -35.8801 Energy for replicate 18 = -35.8376 Energy for replicate 19 = -35.6213 Energy for replicate 20 = -35.4317 Energy for replicate 21 = -34.9259 Energy for replicate 22 = -33.9905 Energy for replicate 23 = -32.3218 Energy for replicate 24 = -30.4850 Energy for replicate 25 = -29.3411 Energy for replicate 26 = -29.8267 Energy for replicate 27 = -31.2547 Energy for replicate 28 = -32.0472 Energy for replicate 29 = -32.7606 Energy for replicate 30 = -33.5716 Energy for replicate 31 = -33.9966 Energy for replicate 32 = -34.1210 Total Energy of replicates = -1109.0158 |
Notice how the energies fluctuate along the pathway. It is probably easier to visualize this if we plot the energies. We can manually extract them an place them in a file: final_pathway_energies.dat.
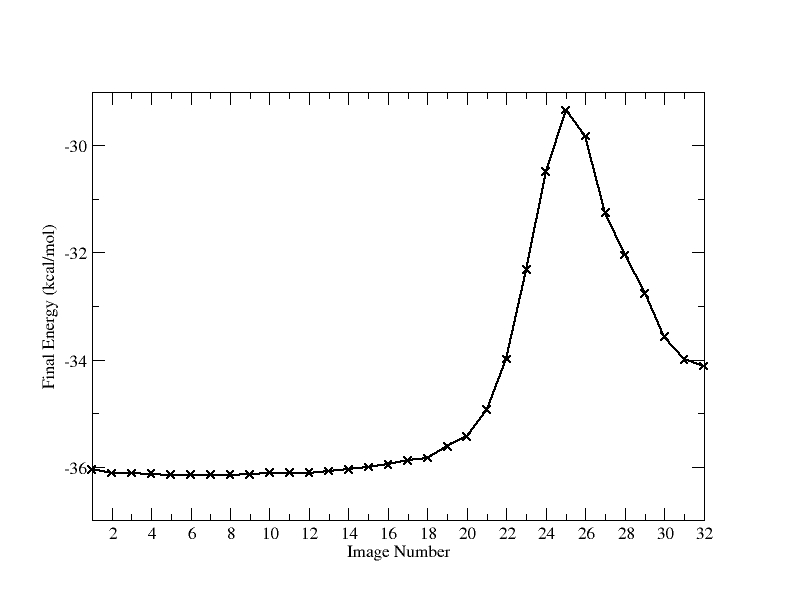
As you can see there is 1 major maxima along this pathway and the overall barrier height is approximately 7 KCal/mol in one direction and 4 KCal/mol in the reverse direction. In reality you would probably want to use at least 32 images here to get a nice smooth curve.


