(Note: These tutorials are meant to provide
illustrative examples of how to use the AMBER software suite to carry out
simulations that can be run on a simple workstation in a reasonable period of
time. They do not necessarily provide the optimal choice of parameters or
methods for the particular application area.)
Copyright Ross Walker 2013
Umbrella Sampling Example
Calculating the PMF for Alanine Dipeptide Phi/Psi Rotation
By Ross Walker & Thomas Steinbrecher
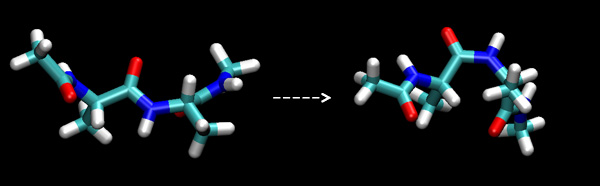
In this tutorial we will learn how to use the AMBER software coupled with the Weighted Histogram Analysis Method (WHAM) of Alan Grossfield to generate potentials of mean force. Often one might want to know what the free energy profile is along a specific reaction coordinate. Such a profile is known as a potential of mean force and it can be very useful for identifying transition states, intermediates as well as the relative stabilities of the end points. At first thought one might think that you could generate a free energy along a specific reaction coordinate by just running an MD simulation and then looking at the probabilities of the states sampled. However, often the energy barrier of interest is many times the size of kbT and so the MD simulation will either remain in the local minimum it started in or cross to different minima but very very rarely sample the transition state. This lack of sampling means that we cannot generate an accurate PMF. Indeed for even small systems we could need in excess of a millisecond of MD to obtain useful sampling. This is obviously not tractable and so a smarter approach must be used.
One such approach is that of umbrella sampling. This works by breaking the reaction coordinate up into a series of windows and then applying a restraint that acts to force the reaction coordinate to remain close to the center of the window. Obviously for ideal sampling the best approach would be to use the free energy profile itself as the umbrella restraint. This would make our energy surface effectively flat and lead to uniform sampling. However, such an approach is not feasible since it requires us to know the shape of the energy surface before we calculate it. Instead what is often done is a series of harmonic potentials are used to constrain the reaction coordinate to various values. From this a series of histograms showing the sampling of the reaction coordinate values can be generated and assuming the windows overlap one can use a weighted histogram analysis method to post process the results to remove the biasing of the restraints and recover the PMF.
In this tutorial we shall use umbrella sampling to generate a PMF for the trans to cis isomerization of alanine tripeptide.
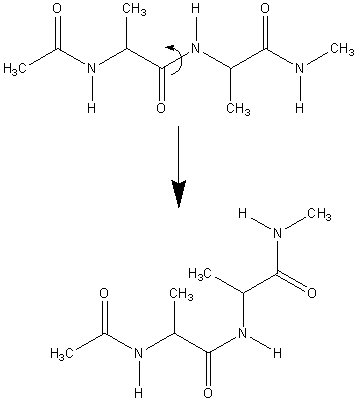
This tutorial consists of three sections:
1) section1 : Creating the initial structure and relaxing it.
2) section2 : Running the Umbrella Sampling Calculations.
3) section3 : Constructing the PMF.
(Note: These tutorials are meant to provide
illustrative examples of how to use the AMBER software suite to carry out
simulations that can be run on a simple workstation in a reasonable period of
time. They do not necessarily provide the optimal choice of parameters or
methods for the particular application area.)
Copyright Ross Walker 2013


