(Note: These tutorials are meant to provide
illustrative examples of how to use the AMBER software suite to carry out
simulations that can be run on a simple workstation in a reasonable period of
time. They do not necessarily provide the optimal choice of parameters or
methods for the particular application area.)
Copyright Ross Walker 2015
Analysis of water thermodynamics using Grid Inhomogeneous Solvation Theory of Factor Xa active site.
By Romelia Salomon, Crystal Nguyen, Steven Ramsey, Jonathan D. Gough, Ross Walker, & Tom Kurtzman
Solvation thermodynamics plays an important role in the free energy of molecular recognition, which makes the analysis of solvation thermodynamics high importance in studies involving protein-drug binding, molecular aggregation, and hydrophobic collapse.
In this tutorial we will learn how to use the AMBER software coupled with the Grid Inhomogeneous Solvation Theory Method (GIST) of Ramsey et.al. 2016, Nguyen et. al. 2012, and Nguyen et. al. 2011, to estimate thermodynamic values for the water molecules occupying the binding pocket of coagulation Factor Xa. Often one might want to estimate changes in hydration which are central to correctly describe biomolecular phenomena such as molecular recognition, drug binding, etc. However it is difficult to estimate precisely the thermodynamic solvation contribution in these phenomena.
GIST calculates thermodynamic values of solvent located within a defined region using the grid discretized inhomogeneous solvation theory formulation. The energy of each voxel represents the full interaction between the water molecules located in that voxels with the solute (usually a protein) and the other waters in the system. The solute-water energy is considered in a full way, ie, the interaction between a water molecule and solute molecules is not divided by two, and is assigned in its totality to the grid box where the water being studied resides. Conversely, water-water energies are halved to prevent double counting.
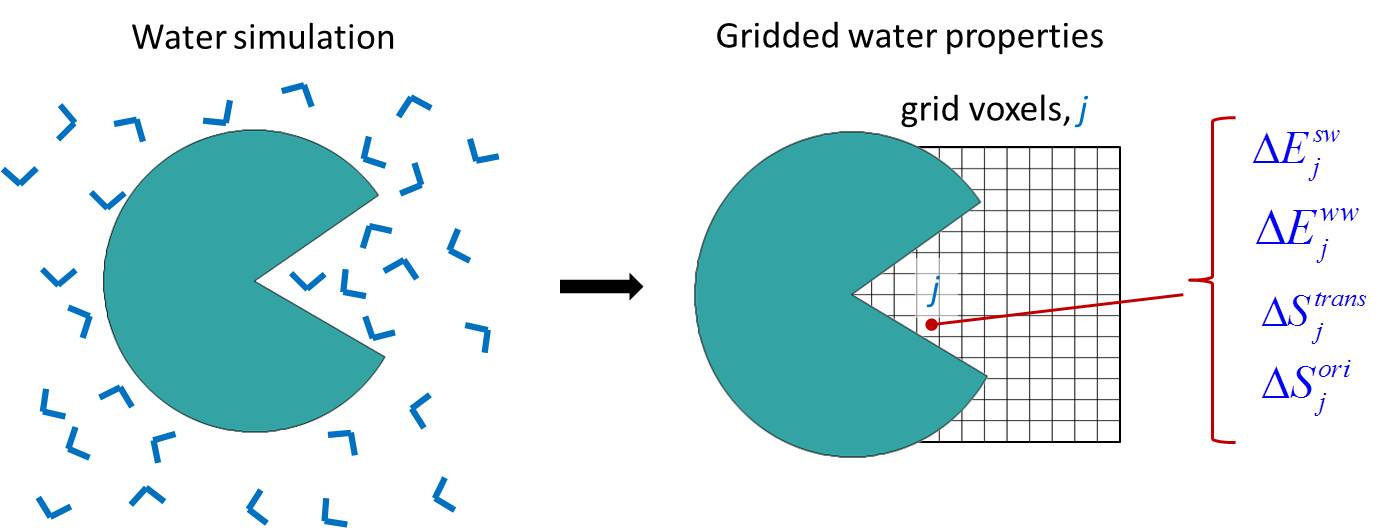
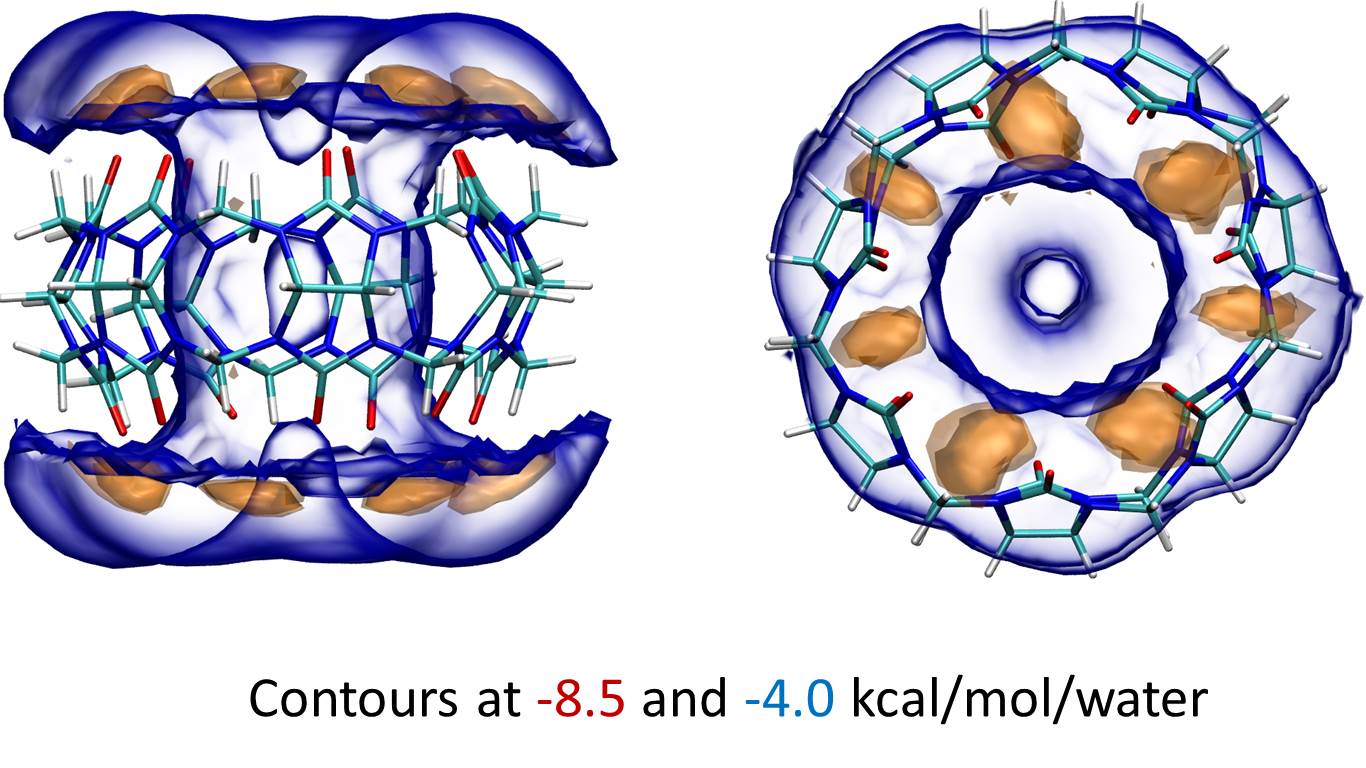
GIST produces quantitative thermodynamic data for each grid box, or voxel. GIST provides the end-user with a detailed map of water thermodynamic and structural quantities in a defined region of interest including: interaction energy of water found in a voxel with all other water molecules or solute molecules, water first order entropic penalty for each voxel, water occupancy within each voxel, etc. This information can be used to decide whether the water at a given location is favorable or not compared to the bulk distribution. The following figure shows the interaction energy contour of a miniature receptor, cucurbit[7]uril, with its surrounding water. The figure shows two favorable water-host energy contours: -8.5 kcal/mol/water in orange and -4.0 kcal/mol/water in blue. The orange contour highlights the more favorable water interaction of the water and carbonyl groups of the host molecule.
The present implementation computes the long range interactions only for the central image, i.e. no periodic copies are considered and no long range methods such as PME are applied. Energy quantities are reported as either density weighted quantities (dens) or averaged per water quantities (norm). At this time, the implementation of GIST in cpptraj supports all three point and four point water molecules available in AMBER, shown here is a select list of said water models:
TIP3P
TIP4P
TIP4PEW
OPC
TIP3PFW (flexible water model)
SPCE
SPCFW (flexible water model)
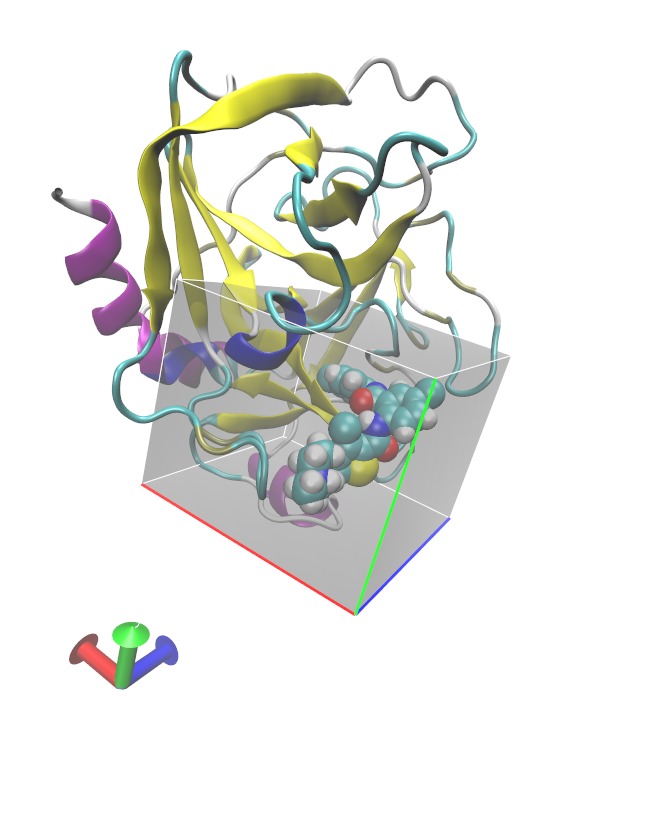
In this tutorial we shall use GIST to evaluate the thermodynamic contributions of hydration in the active site of Factor Xa. (The colored axes show the edges of the grid used to define GIST active space for this calculation, we have placed a ligand in the pocket for visual aid.)
This tutorial consists of four sections:
1) section1 : Creating the initial structure and relaxing it.
2) section2 : Running the MD calculations in Amber.
3) section3 : Use GIST in cpptraj to estimate thermodynamic values of hydration.
4) section4 : Analysis of water characteristics through GISTPP application to GIST output files
(Note: These tutorials are meant to provide
illustrative examples of how to use the AMBER software suite to carry out
simulations that can be run on a simple workstation in a reasonable period of
time. They do not necessarily provide the optimal choice of parameters or
methods for the particular application area.)
Copyright Ross Walker 2015


